import numpy as np
from scipy.stats import multivariate_normal
import numpy as np
import matplotlib.pyplot as plt
from scipy.stats import multivariate_normal
def initialize_parameters(data, num_components):
"""
Initializes the parameters: mixture weights, means, and covariances.
"""
num_features = data.shape[1]
weights = np.ones(num_components) / num_components
means = data[np.random.choice(data.shape[0], num_components, False)]
covariances = np.array([np.cov(data.T) for _ in range(num_components)])
return weights, means, covariances
def e_step(data, weights, means, covariances):
"""
The E-step of the EM algorithm.
"""
num_samples = data.shape[0]
num_components = len(weights)
responsibilities = np.zeros((num_samples, num_components))
for i in range(num_components):
responsibilities[:, i] = weights[i] * multivariate_normal.pdf(data, means[i], covariances[i])
responsibilities /= responsibilities.sum(axis=1, keepdims=True)
return responsibilities
def m_step(data, responsibilities):
"""
The M-step of the EM algorithm.
"""
num_samples, num_features = data.shape
num_components = responsibilities.shape[1]
weights = responsibilities.sum(axis=0) / num_samples
means = np.dot(responsibilities.T, data) / responsibilities.sum(axis=0)[:, np.newaxis]
covariances = np.zeros((num_components, num_features, num_features))
for i in range(num_components):
diff = data - means[i]
covariances[i] = np.dot(responsibilities[:, i] * diff.T, diff) / responsibilities[:, i].sum()
return weights, means, covariances
def gmm_em(data, num_components, num_iterations):
"""
EM algorithm for a Gaussian Mixture Model.
"""
weights, means, covariances = initialize_parameters(data, num_components)
for _ in range(num_iterations):
responsibilities = e_step(data, weights, means, covariances)
weights, means, covariances = m_step(data, responsibilities)
return weights, means, covariances
# Example usage
data = np.random.randn(100, 2)+10 * np.random.randn(100, 2) # Replace with your dataset
def plot_data(data):
plt.figure(figsize=(10, 6))
plt.scatter(data[:, 0], data[:, 1], s=30, alpha=0.5, label="data points sampled from unknown $p_{data}$")
colors = ['r', 'g', 'b', 'c', 'm', 'y', 'k']
x, y = np.mgrid[np.min(data[:,0])-1:np.max(data[:,0])+1:.01, np.min(data[:,1])-1:np.max(data[:,1])+1:.01]
pos = np.dstack((x, y))
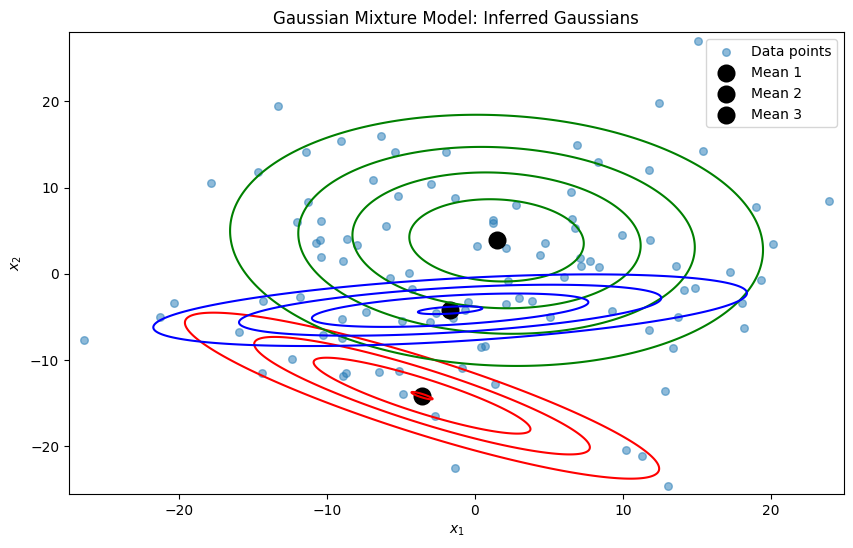
# for i, (mean, cov) in enumerate(zip(means, covariances)):
# rv = multivariate_normal(mean, cov)
# plt.contour(x, y, rv.pdf(pos), colors=colors[i], levels=5)
# plt.scatter(mean[0], mean[1], marker='o', color='k', s=100, lw=3, label=f"Mean {i+1}")
plt.title("Raw data points")
plt.xlabel("$x_1$")
plt.ylabel("$x_2$")
plt.legend()
plt.show()
plot_data(data)