import math
import numpy as np
import torch
import torch.nn as nn
from torch.utils.data import TensorDataset, DataLoader
import matplotlib.pyplot as plt
torch.manual_seed(7)
np.random.seed(7)
device = "cpu"
def train_test_split(X, y, test_size=0.3, shuffle=True, seed=0):
n = X.shape[0]
idx = np.arange(n)
if shuffle:
rng = np.random.default_rng(seed)
rng.shuffle(idx)
n_test = int(round(test_size * n))
test_idx = idx[:n_test]
train_idx = idx[n_test:]
return X[train_idx], X[test_idx], y[train_idx], y[test_idx]
def standardize_fit(X):
mu = X.mean(axis=0, keepdims=True)
std = X.std(axis=0, keepdims=True) + 1e-8
return mu, std
def standardize_transform(X, mu, std):
return (X - mu) / std
def auc_roc(scores, labels):
scores = np.asarray(scores).ravel()
labels = np.asarray(labels).astype(int).ravel()
order = np.argsort(-scores, kind="mergesort")
y = labels[order]
P = y.sum()
N = len(y) - P
if P == 0 or N == 0:
return float("nan")
tps = np.cumsum(y)
fps = np.cumsum(1 - y)
tpr = tps / P
fpr = fps / N
return float(np.trapz(tpr, fpr))Logistic Regression with SGD on a Gaussian GMM Dataset + PCA
Objective. Generate a synthetic Gaussian Mixture Model (GMM) binary classification dataset where only a low‑dimensional subspace is informative and the remaining dimensions are noisy.
Train logistic regression with SGD in PyTorch with and without PCA, and compare performance.
What students should learn - How redundant/noisy features degrade a linear classifier trained with SGD. - How PCA (unsupervised) can improve performance by denoising and reducing dimensionality. - How to visualize explained variance and decision boundaries in the top PCs.
Setup
Generate a GMM dataset (binary) with noisy dimensions
def make_gmm_binary(n_per_class=1000, d_total=20, d_informative=2, class_sep=2.5, noise_std=1.0, seed=0):
rng = np.random.default_rng(seed)
sep = class_sep
mus0 = np.array([[-sep, 0.0], [0.0, -sep]])
mus1 = np.array([[sep, 0.0], [0.0, sep]])
cov_inf = np.array([[1.0, 0.3], [0.3, 1.0]])
def sample_class(mus, n):
z = rng.integers(0, len(mus), size=n)
X2 = np.stack([rng.multivariate_normal(mus[k], cov_inf) for k in z], axis=0)
return X2
X0_inf = sample_class(mus0, n_per_class)
X1_inf = sample_class(mus1, n_per_class)
d_noise = max(0, d_total - d_informative)
if d_noise > 0:
X0_noise = rng.normal(0.0, noise_std, size=(n_per_class, d_noise))
X1_noise = rng.normal(0.0, noise_std, size=(n_per_class, d_noise))
X0 = np.concatenate([X0_inf, X0_noise], axis=1)
X1 = np.concatenate([X1_inf, X1_noise], axis=1)
else:
X0 = X0_inf
X1 = X1_inf
X = np.vstack([X0, X1]).astype(np.float32)
y = np.concatenate([np.zeros(n_per_class), np.ones(n_per_class)]).astype(np.int64)
Q, _ = np.linalg.qr(rng.normal(size=(d_total, d_total)))
X = (X @ Q).astype(np.float32)
return X, y# X, y = make_gmm_binary(n_per_class=1200, d_total=20, d_informative=2, class_sep=2.5, noise_std=1.5, seed=42)
X, y = make_gmm_binary(n_per_class=200, d_total=20, d_informative=2, class_sep=2.5, noise_std=1.0, seed=42)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, shuffle=True, seed=123)
mu, std = standardize_fit(X_train)
Xtr = standardize_transform(X_train, mu, std)
Xte = standardize_transform(X_test, mu, std)
X.shape, Xtr.shape, Xte.shape, y_train.shape, y_test.shape((400, 20), (280, 20), (120, 20), (280,), (120,))PCA (Torch SVD)
def pca_fit_torch(X_np):
Xc = X_np - X_np.mean(axis=0, keepdims=True)
Xc_t = torch.from_numpy(Xc)
U, S, Vh = torch.linalg.svd(Xc_t, full_matrices=False)
n = X_np.shape[0]
ev = (S**2) / (n - 1)
evr = ev / ev.sum()
components = Vh.numpy()
return components, evr.numpy()
def pca_transform(X_np, components, k):
Xc = X_np - X_np.mean(axis=0, keepdims=True)
W = components[:k].T
return (Xc @ W).astype(np.float32)
components, evr = pca_fit_torch(Xtr.copy())
explained = np.cumsum(evr)
k95 = int(np.searchsorted(explained, 0.95) + 1)
k_fixed = 2
print("Explained variance ratio (first 10):", evr[:10])
print("k@95% =", k95)Explained variance ratio (first 10): [0.14723612 0.12738939 0.06149405 0.05959329 0.05904397 0.05309147
0.05024018 0.04745107 0.04639223 0.04420933]
k@95% = 18Plot: cumulative explained variance
plt.figure(figsize=(6, 4))
plt.plot(np.arange(1, len(evr) + 1), np.cumsum(evr), marker="o")
plt.xlabel("number of PCs")
plt.ylabel("cumulative explained variance")
plt.title("PCA cumulative explained variance on training set")
plt.show()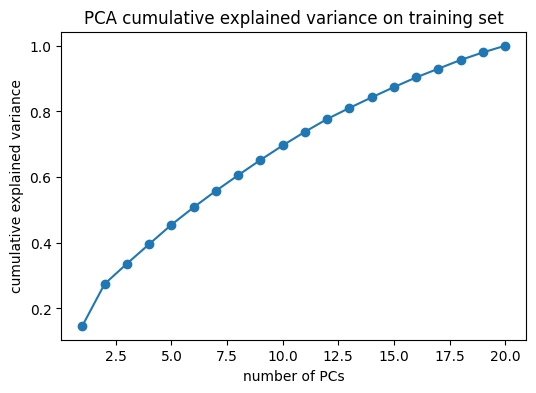
Logistic regression (PyTorch, SGD)
class LogisticReg(nn.Module):
def __init__(self, in_dim):
super().__init__()
self.lin = nn.Linear(in_dim, 1)
def forward(self, x):
return self.lin(x).squeeze(-1)
def auc_roc(scores, labels):
scores = np.asarray(scores).ravel()
labels = np.asarray(labels).astype(int).ravel()
order = np.argsort(-scores, kind="mergesort")
y = labels[order]
P = y.sum()
N = len(y) - P
if P == 0 or N == 0:
return float("nan")
tps = np.cumsum(y)
fps = np.cumsum(1 - y)
tpr = tps / P
fpr = fps / N
return float(np.trapz(tpr, fpr))
def train_logreg_sgd(Xtr_np, ytr_np, Xte_np, yte_np, lr=5e-3, epochs=300, batch_size=128, l2=1e-4):
Xtr_t = torch.from_numpy(Xtr_np.astype(np.float32))
ytr_t = torch.from_numpy(ytr_np.astype(np.float32))
Xte_t = torch.from_numpy(Xte_np.astype(np.float32))
yte_t = torch.from_numpy(yte_np.astype(np.float32))
model = LogisticReg(in_dim=Xtr_t.shape[1])
opt = torch.optim.SGD(model.parameters(), lr=lr, momentum=0.9, weight_decay=l2)
loss_fn = nn.BCEWithLogitsLoss()
ds = TensorDataset(Xtr_t, ytr_t)
dl = DataLoader(ds, batch_size=batch_size, shuffle=True)
model.train()
for ep in range(1, epochs + 1):
running = 0.0
for xb, yb in dl:
logits = model(xb)
loss = loss_fn(logits, yb)
opt.zero_grad()
loss.backward()
opt.step()
running += loss.item() * xb.size(0)
if ep % 50 == 0:
print(f"epoch {ep:4d} | loss: {running / len(ds):.4f}")
model.eval()
with torch.no_grad():
logits_tr = model(Xtr_t).numpy()
logits_te = model(Xte_t).numpy()
prob_tr = 1.0 / (1.0 + np.exp(-logits_tr))
prob_te = 1.0 / (1.0 + np.exp(-logits_te))
pred_te = (prob_te >= 0.5).astype(int)
acc_te = (pred_te == yte_np).mean()
auc_te = auc_roc(prob_te, yte_np)
return model, acc_te, auc_te, prob_te, logits_teTrain & evaluate: baseline vs PCA
model_raw, acc_raw, auc_raw, prob_raw, logit_raw = train_logreg_sgd(
Xtr, y_train, Xte, y_test, lr=5e-3, epochs=300, batch_size=128, l2=1e-4
)
print(f"Baseline (no PCA): acc={acc_raw:.3f}, AUC={auc_raw:.3f}")
Xtr_pca95 = pca_transform(Xtr, components, k95)
Xte_pca95 = pca_transform(Xte, components, k95)
model_pca95, acc_pca95, auc_pca95, prob_pca95, _ = train_logreg_sgd(
Xtr_pca95, y_train, Xte_pca95, y_test, lr=5e-3, epochs=300, batch_size=128, l2=1e-4
)
print(f"PCA (k@95%={k95}): acc={acc_pca95:.3f}, AUC={auc_pca95:.3f}")
Xtr_pca2 = pca_transform(Xtr, components, 2)
Xte_pca2 = pca_transform(Xte, components, 2)
model_pca2, acc_pca2, auc_pca2, prob_pca2, _ = train_logreg_sgd(
Xtr_pca2, y_train, Xte_pca2, y_test, lr=5e-3, epochs=300, batch_size=128, l2=1e-4
)
print(f"PCA (k=2): acc={acc_pca2:.3f}, AUC={auc_pca2:.3f}")epoch 50 | loss: 0.1858
epoch 100 | loss: 0.1544
epoch 150 | loss: 0.1423
epoch 200 | loss: 0.1353
epoch 250 | loss: 0.1308
epoch 300 | loss: 0.1278
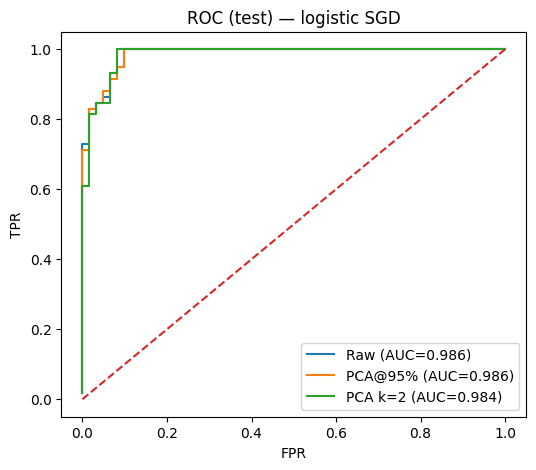
Baseline (no PCA): acc=0.950, AUC=0.986
epoch 50 | loss: 0.1850
epoch 100 | loss: 0.1537
epoch 150 | loss: 0.1422
epoch 200 | loss: 0.1358
epoch 250 | loss: 0.1317
epoch 300 | loss: 0.1289
PCA (k@95%=18): acc=0.933, AUC=0.986
epoch 50 | loss: 0.1929
epoch 100 | loss: 0.1670
epoch 150 | loss: 0.1584
epoch 200 | loss: 0.1545
epoch 250 | loss: 0.1523
epoch 300 | loss: 0.1506
PCA (k=2): acc=0.933, AUC=0.984Plot: decision boundary in 2D PCA space
xx, yy = np.meshgrid(
np.linspace(Xtr_pca2[:, 0].min() - 1.0, Xtr_pca2[:, 0].max() + 1.0, 200),
np.linspace(Xtr_pca2[:, 1].min() - 1.0, Xtr_pca2[:, 1].max() + 1.0, 200),
)
grid = np.stack([xx.ravel(), yy.ravel()], axis=1).astype(np.float32)
with torch.no_grad():
logits_grid = model_pca2.lin(torch.from_numpy(grid)).numpy().reshape(xx.shape)
probs_grid = 1.0 / (1.0 + np.exp(-logits_grid))
plt.figure(figsize=(6, 5))
plt.contourf(xx, yy, probs_grid, levels=20, alpha=0.3)
plt.scatter(Xtr_pca2[:, 0], Xtr_pca2[:, 1], s=10, c=y_train, alpha=0.6)
plt.title("Decision surface (logistic on top-2 PCs)")
plt.xlabel("PC1")
plt.ylabel("PC2")
plt.show()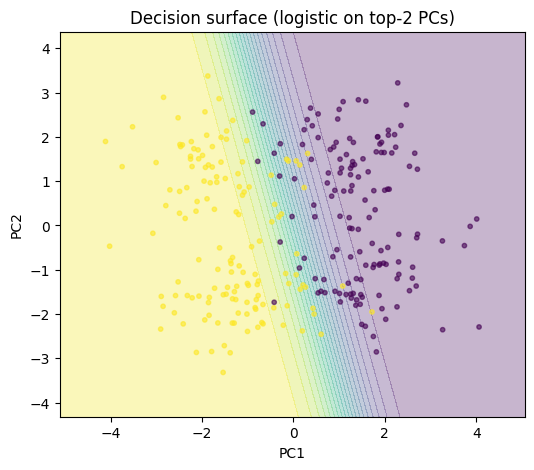
Plot: ROC curves (test set) — raw vs PCA
def roc_curve(scores, labels):
scores = np.asarray(scores).ravel()
labels = np.asarray(labels).astype(int).ravel()
order = np.argsort(-scores, kind="mergesort")
y = labels[order]
P = y.sum()
N = len(y) - P
tps = np.cumsum(y)
fps = np.cumsum(1 - y)
tpr = tps / (P + 1e-12)
fpr = fps / (N + 1e-12)
return fpr, tpr
fpr_raw, tpr_raw = roc_curve(prob_raw, y_test)
fpr_95, tpr_95 = roc_curve(prob_pca95, y_test)
fpr_2, tpr_2 = roc_curve(prob_pca2, y_test)
plt.figure(figsize=(6, 5))
plt.plot(fpr_raw, tpr_raw, label=f"Raw (AUC={auc_raw:.3f})")
plt.plot(fpr_95, tpr_95, label=f"PCA@95% (AUC={auc_pca95:.3f})")
plt.plot(fpr_2, tpr_2, label=f"PCA k=2 (AUC={auc_pca2:.3f})")
plt.plot([0, 1], [0, 1], linestyle="--")
plt.xlabel("FPR")
plt.ylabel("TPR")
plt.title("ROC (test) — logistic SGD")
plt.legend()
plt.show()