import torch
import math
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
torch.manual_seed(0)
print("PyTorch:", torch.__version__)PyTorch: 2.7.1+cu126Goal. Use Monte Carlo experiments to show that, for a fixed number of samples (n), data become effectively “sparser” as the input dimension (d) grows.
Setup. For each (d {1,2,5,10,20,50,100}), draw (n) i.i.d. points (X_1,,X_n ([0,1]^d)).
Tasks. 1. Nearest-neighbor distance grows. Compute the nearest-neighbor (NN) distance for each point and report the mean NN distance as a function of (d). Show that it increases with (d). 2. Distance concentration. Compute all pairwise Euclidean distances and report the coefficient of variation (std/mean). Show that the relative spread of distances shrinks with (d) (concentration of measure), making points nearly equidistant—hence no “close” neighbors. 3. Vanishing local mass. Fix a small radius (r>0) (e.g., (r=0.10)). Let (c=(0.5,,0.5)) be the hypercube center. Estimate the fraction of points within the ball (B(c,r)). Show it decreases with (d). 4. Local neighborhood empties. For a fixed small radius (r) (e.g., (r=0.15)), count how many neighbors each point has within that radius and report the average count vs (d). Show it decreases.
Deliverables. - A table with the metrics above for each (d). - Plots of (i) mean NN distance, (ii) coefficient of variation of pairwise distances, (iii) fraction inside (B(c,r)), (iv) average neighbor count within (r) — all vs (d).
Interpretation. Relate these observations to the curse of dimensionality. Explain how increased NN distances and distance concentration imply that models relying on local neighborhoods (e.g., kNN, kernels) struggle in high dimensions, and why uncertainty estimates (e.g., MLE variance) tend to inflate in sparse regions.
def pairwise_distances(x: torch.Tensor) -> torch.Tensor:
"""Compute Euclidean pairwise distances using torch.cdist.
Args:
x: (n, d) tensor
Returns:
(n, n) distance matrix
"""
return torch.cdist(x, x)
def nearest_neighbor_distances(D: torch.Tensor) -> torch.Tensor:
"""Nearest-neighbor distance per point from a distance matrix.
Args:
D: (n, n) distance matrix
Returns:
(n,) tensor of NN distances
"""
D_masked = D + torch.eye(D.size(0)) * 1e9
nn_dist, _ = torch.min(D_masked, dim=1)
return nn_distrows = []
for d in dims:
# 1) Sample points in [0,1]^d
X = torch.rand(n, d)
# 2) Pairwise distances and NN stats
D = pairwise_distances(X)
tri = torch.triu_indices(n, n, offset=1)
pdists = D[tri[0], tri[1]]
pair_mean = pdists.mean().item()
pair_std = pdists.std(unbiased=False).item()
pair_cv = pair_std / (pair_mean + 1e-12)
nn = nearest_neighbor_distances(D)
mean_nn = nn.mean().item()
median_nn = nn.median().item()
# 3) Fraction inside ball around center
center = torch.full((d,), 0.5)
dist_to_center = torch.linalg.norm(X - center, dim=1)
frac_in_ball = (dist_to_center <= r_center).float().mean().item()
# 4) Average neighbors within fixed radius
within = (D <= r_neighbors).float()
within.fill_diagonal_(0.0)
avg_neighbors = within.sum(dim=1).mean().item()
rows.append(
{
"d": d,
"mean_NN": mean_nn,
"median_NN": median_nn,
"pair_mean": pair_mean,
"pair_std": pair_std,
"pair_CV": pair_cv,
f"frac_in_ball(r={r_center})": frac_in_ball,
f"avg_neighbors(r={r_neighbors})": avg_neighbors,
"n": n,
}
)
df = pd.DataFrame(rows)
df| d | mean_NN | median_NN | pair_mean | pair_std | pair_CV | frac_in_ball(r=0.1) | avg_neighbors(r=0.15) | n | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 0.000402 | 0.000299 | 0.337809 | 0.238667 | 0.706515 | 0.199167 | 328.850006 | 1200 |
| 1 | 2 | 0.014385 | 0.013267 | 0.514249 | 0.244681 | 0.475803 | 0.031667 | 73.551666 | 1200 |
| 2 | 5 | 0.171974 | 0.171206 | 0.876517 | 0.248289 | 0.283267 | 0.000000 | 0.390000 | 1200 |
| 3 | 10 | 0.492865 | 0.494772 | 1.269582 | 0.245546 | 0.193407 | 0.000000 | 0.000000 | 1200 |
| 4 | 20 | 1.017114 | 1.018217 | 1.799921 | 0.243045 | 0.135031 | 0.000000 | 0.000000 | 1200 |
| 5 | 50 | 2.111619 | 2.115882 | 2.876143 | 0.241308 | 0.083900 | 0.000000 | 0.000000 | 1200 |
| 6 | 100 | 3.329890 | 3.332430 | 4.081142 | 0.239164 | 0.058602 | 0.000000 | 0.000000 | 1200 |
# 1) Mean NN distance vs d
plt.figure(figsize=(6, 4))
plt.plot(df["d"], df["mean_NN"], marker="o")
plt.xlabel("dimension d")
plt.ylabel("mean nearest-neighbor distance")
plt.title("Mean NN distance grows with dimension")
plt.show()
# 2) Coefficient of variation of pairwise distances vs d
plt.figure(figsize=(6, 4))
plt.plot(df["d"], df["pair_CV"], marker="o")
plt.xlabel("dimension d")
plt.ylabel("coeff. of variation of pairwise distances")
plt.title("Distance concentration (CV ↓) as dimension increases")
plt.show()
# 3) Fraction inside ball around center vs d
plt.figure(figsize=(6, 4))
plt.plot(df["d"], df[f"frac_in_ball(r={r_center})"], marker="o")
plt.xlabel("dimension d")
plt.ylabel(f"fraction inside ball (r={r_center}) around center")
plt.title("Local mass around center vanishes with dimension")
plt.show()
# 4) Average #neighbors within fixed radius vs d
plt.figure(figsize=(6, 4))
plt.plot(df["d"], df[f"avg_neighbors(r={r_neighbors})"], marker="o")
plt.xlabel("dimension d")
plt.ylabel(f"avg neighbors within r={r_neighbors}")
plt.title("Local neighborhoods empty in higher dimensions")
plt.show()
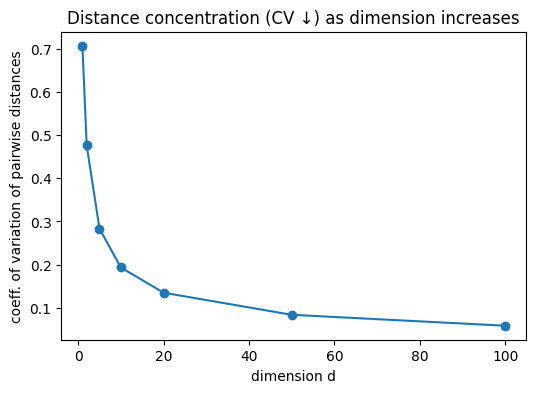
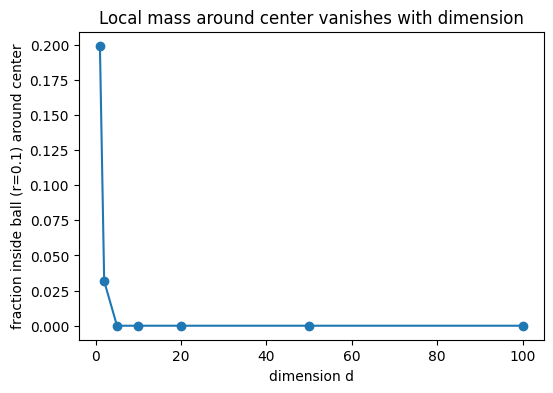
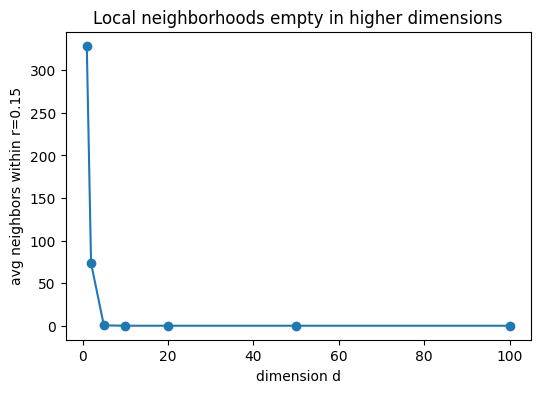
These effects illustrate the curse of dimensionality: methods reliant on local neighborhoods (kNN, kernels) become data-hungry; likewise, uncertainty estimates (e.g., MLE variance) inflate away from the rare dense pockets of data.